Support Vector Machines (svm)¶
The module for Support Vector Machine (SVM) classification is based on the popular LibSVM and LIBLINEAR libraries. It provides several learning algorithms:
SVMLearner, a general SVM learner;
- SVMLearnerEasy, which is similar to the svm-easy.py script
from the LibSVM distribution and helps with the data normalization and parameter tuning;
- LinearSVMLearner, a fast learner useful for data sets with a large
number of features.
SVM learners (from LibSVM)¶
SVMLearner uses the standard LibSVM learner. It supports several built-in kernel types and user-defined kernels functions written in Python. The kernel type is denoted by constants Linear, Polynomial, RBF, Sigmoid and Custom defined in Orange.classification.svm.kernels. A custom kernel function must accept two data instances and return a float. See Kernel wrappers for examples.
The class also supports several types of optimization: C_SVC, Nu_SVC (default), OneClass, Epsilon_SVR and Nu_SVR (defined in Orange.classification.svm.SVMLearner).
Class SVMLearner works on non-sparse data and SVMLearnerSparse class works on sparse data sets, for instance data from the basket format).
- class Orange.classification.svm.SVMLearner(svm_type=Nu_SVC, kernel_type=RBF, kernel_func=None, C=1, nu=0.5, p=0.1, gamma=0.0, degree=3, coef0=0, shrinking=True, probability=True, verbose=False, cache_size=200, eps=0.001, normalization=True, weight=, []**kwargs)¶
Parameters: - svm_type (SVMLearner.SVMType) – the SVM type
- kernel_type (SVMLearner.Kernel) – the kernel type
- degree (int) – kernel parameter (only for Polynomial)
- gamma (float) – kernel parameter; if 0, it is set to 1.0/#features (for Polynomial, RBF and Sigmoid)
- coef0 (int) – kernel parameter (for Polynomial and Sigmoid)
- kernel_func (callable object) – kernel function if kernel_type is kernels.Custom
- C (float) – C parameter (for C_SVC, Epsilon_SVR and Nu_SVR)
- nu (float) – Nu parameter (for Nu_SVC, Nu_SVR and OneClass)
- p (float) – epsilon parameter (for Epsilon_SVR)
- cache_size (int) – cache memory size in MB
- eps (float) – tolerance of termination criterion
- probability (bool) – build a probability model
- shrinking (bool) – use shrinking heuristics
- normalization (bool) – normalize the input data prior to learning into range [0..1] and replace discrete features with indicator columns one for each value of the feature using DomainContinuizer class (default True)
- weight (list) – a list of class weights
- verbose (bool) – If True show training progress (default is False).
Example:
>>> import Orange >>> from Orange.classification import svm >>> from Orange.evaluation import testing, scoring >>> data = Orange.data.Table("vehicle.tab") >>> learner = svm.SVMLearner() >>> results = testing.cross_validation([learner], data, folds=5) >>> print "CA: %.2f" % scoring.CA(results)[0] CA: 0.79 >>> print "AUC: %.2f" % scoring.AUC(results)[0] AUC: 0.95
- __call__(data, weight=0)¶
Construct a SVM classifier
Parameters: - table (Orange.data.Table) – data with continuous features
- weight – ignored (required due to base class signature);
- static max_nu(data)¶
Return the maximum nu parameter for the given data table for Nu_SVC learning.
Parameters: data (Orange.data.Table) – Data with discrete class variable
- tune_parameters(data, parameters=None, folds=5, verbose=0, progress_callback=None)¶
Tune the parameters on the given data using internal cross validation.
Parameters: - data (Orange.data.Table) – data for parameter tuning
- parameters (list of strings) – names of parameters to tune (default: [“nu”, “C”, “gamma”])
- folds (int) – number of folds for internal cross validation
- verbose (bool) – set verbose output
- progress_callback (callback function) – callback function for reporting progress
Here is example of tuning the gamma parameter using 3-fold cross validation.
svm = Orange.classification.svm.SVMLearner() svm.tune_parameters(table, parameters=["gamma"], folds=3)
- class Orange.classification.svm.SVMLearnerSparse(**kwds)¶
Bases: Orange.classification.svm.SVMLearner
A SVMLearner that learns from data stored in meta attributes. Meta attributes do not need to be registered with the data set domain, or present in all data instances.
- class Orange.classification.svm.SVMLearnerEasy(folds=4, verbose=0, **kwargs)¶
Bases: Orange.classification.svm.SVMLearner
A class derived from SVMLearner that automatically scales the data and performs parameter optimization using SVMLearner.tune_parameters(). The procedure is similar to that implemented in easy.py script from the LibSVM package.
- __init__(folds=4, verbose=0, **kwargs)¶
Parameters: kwargs is passed to SVMLearner
The example below compares the performances of SVMLearnerEasy with automatic data preprocessing and parameter tuning and SVMLearner with the default nu and gamma:
from Orange import data
from Orange.classification import svm
vehicle = data.Table("vehicle.tab")
svm_easy = svm.SVMLearnerEasy(name="svm easy", folds=3)
svm_normal = svm.SVMLearner(name="svm")
learners = [svm_easy, svm_normal]
from Orange.evaluation import testing, scoring
results = testing.proportion_test(learners, vehicle, times=1)
print "Name CA AUC"
for learner,CA,AUC in zip(learners, scoring.CA(results), scoring.AUC(results)):
print "%-8s %.2f %.2f" % (learner.name, CA, AUC)
Linear SVM learners (from LIBLINEAR)¶
Linear SVM learners are more suitable for large scale problems since they are significantly faster then SVMLearner and its subclasses. A down side is that they support only a linear kernel and can not estimate probabilities.
- class Orange.classification.svm.LinearSVMLearner(solver_type=L2R_L2Loss_SVC_Dual, C=1, eps=0.01, bias=1, normalization=True, multinomial_treatment=NValues, **kwargs)¶
Train a linear SVM model.
- __init__(solver_type=L2R_L2Loss_SVC_Dual, C=1, eps=0.01, bias=1, normalization=True, multinomial_treatment=NValues, **kwargs)¶
Parameters: - solver_type –
One of the following class constants: L2R_L2LOSS_DUAL, L2R_L2LOSS, L2R_L1LOSS_DUAL, L1R_L2LOSS
The first part (L2R or L1R) is the regularization term on the weight vector (squared or absolute norm respectively), the L1LOSS or L2LOSS indicate absolute or squared loss function DUAL means the optimization problem is solved in the dual space (for more information see the documentation on LIBLINEAR).
- C (float) – Regularization parameter (default 1.0)
- eps (float) – Stopping criteria (default 0.01)
- bias (float) – If non negative then each instance is appended a constant bias term (default 1.0).
- normalization (bool) – Normalize the input data into range [0..1] prior to learning (default True)
- multinomial_treatment (int) – Defines how to handle multinomial features for learning. It can be one of the DomainContinuizer multinomial_treatment constants (default: DomainContinuizer.NValues).
New in version 2.6.1: Added multinomial_treatment
Note
By default if the training data contains discrete features they are replaced by indicator columns one for each value of the feature regardless of the value of normalization. This is different then in SVMLearner where this is done only if normalization is True.
Example
>>> linear_svm = LinearSVMLearner( ... solver_type=LinearSVMLearner.L1R_L2LOSS, ... C=2.0) ...
- solver_type –
- class Orange.classification.svm.MultiClassSVMLearner(C=1, eps=0.01, bias=1, normalization=True, multinomial_treatment=NValues, **kwargs)¶
Multi-class SVM (Crammer and Singer) from the LIBLINEAR library.
- __init__(C=1, eps=0.01, bias=1, normalization=True, multinomial_treatment=NValues, **kwargs)¶
Parameters: - C (float) – Regularization parameter (default 1.0)
- eps (float) – Stopping criteria (default 0.01)
- bias (float) – If non negative then each instance is appended a constant bias term (default 1.0).
- normalization (bool) – Normalize the input data prior to learning (default True)
- multinomial_treatment (int) – Defines how to handle multinomial features for learning. It can be one of the DomainContinuizer multinomial_treatment constants (default: DomainContinuizer.NValues).
New in version 2.6.1: Added multinomial_treatment
- class Orange.classification.svm.LinearClassifier¶
The classifier returned by LIBLINEAR based learners.
- weights¶
A 2 dim table of computed feature weights of the classifier, one for each one vs. rest underlying binary classifier (i.e. classifier.weights[i] contains the i’th class vs. rest binary classifier weights. If bias > 0 then the bias weight term is appended as the last element of the weight vector.
- bias¶
The bias parameter as passed to the learner.
SVM Based feature selection and scoring¶
- class Orange.classification.svm.RFE(learner=None)¶
Iterative feature elimination based on weights computed by a linear SVM.
Example:
>>> table = Orange.data.Table("promoters.tab") >>> svm_l = Orange.classification.svm.SVMLearner( ... kernel_type=Orange.classification.svm.kernels.Linear) ... >>> rfe = Orange.classification.svm.RFE(learner=svm_l) >>> data_with_subset_of_features = rfe(table, 10) >>> data_with_subset_of_features.domain [p-45, p-36, p-35, p-34, p-33, p-31, p-18, p-12, p-10, p-04, y]
- __call__(data, num_selected=20, progress_callback=None)¶
Return a new dataset with only num_selected best scoring attributes.
Parameters: - data (Orange.data.Table) – Data
- num_selected (int) – number of features to preserve
- __init__(learner=None)¶
Parameters: learner (LinearSVMLearner or SVMLearner with linear kernel) – A linear svm learner for use for scoring (this learner is passed to ScoreSVMWeights) See also
- class Orange.classification.svm.ScoreSVMWeights(learner=None, **kwargs)¶
Bases: Orange.feature.scoring.Score
Score a feature using squared weights of a linear SVM model.
Example:
>>> table = Orange.data.Table("vehicle.tab") >>> score = Orange.classification.svm.ScoreSVMWeights() >>> svm_scores = [(score(f, table), f) for f in table.domain.features] >>> for feature_score, feature in sorted(svm_scores, reverse=True): ... print "%-35s: %.1f" % (feature.name, feature_score) pr.axis aspect ratio : 44.3 kurtosis about major axis : 42.6 max.length rectangularity : 39.4 radius ratio : 28.7 ...
- __init__(learner=None, **kwargs)¶
Parameters: learner (Orange.core.LinearLearner) – Learner used for weight estimation (by default LinearSVMLearner(solver_type=L2R_L2LOSS_DUAL, C=1.0) will be used for classification problems and SVMLearner(svm_type=Epsilon_SVR, kernel_type=Linear, C=1.0, p=0.25) for regression problems).
Utility functions¶
- static svm.max_nu(data)¶
Return the maximum nu parameter for the given data table for Nu_SVC learning.
Parameters: data (Orange.data.Table) – Data with discrete class variable
- static svm.get_linear_svm_weights(classifier, sum=True)¶
Extract attribute weights from the linear SVM classifier.
For multi class classification, the result depends on the argument sum. If True (default) the function computes the squared sum of the weights over all binary one vs. one classifiers. If sum is False it returns a list of weights for each individual binary classifier (in the order of [class1 vs class2, class1 vs class3 ... class2 vs class3 ...]).
- static svm.table_to_svm_format(data, file)¶
Save Orange.data.Table to a format used by LibSVM.
Parameters: - data (Orange.data.Table) – Data
- file (file) – file pointer
The following example shows how to get linear SVM weights:
from Orange import data
from Orange.classification import svm
brown = data.Table("brown-selected")
classifier = svm.SVMLearner(brown,
kernel_type=svm.kernels.Linear,
normalization=False,
eps=1e-9)
weights = svm.get_linear_svm_weights(classifier)
print sorted("%.4f" % w for w in weights.values())
import pylab as plt
plt.hist(weights.values())
Kernel wrappers¶
Kernel wrappers are helper classes for building custom kernels for use with SVMLearner and subclasses. They take and transform one or two Python functions (attributes wrapped or wrapped1 and wrapped2). The function must be a positive definite kernel that takes two arguments of type Orange.data.Instance and returns a float.
- class Orange.classification.svm.kernels.KernelWrapper(wrapped)¶
A base class for kernel function wrappers.
Parameters: wrapped – a kernel function to wrap
- class Orange.classification.svm.kernels.DualKernelWrapper(wrapped1, wrapped2)¶
A base class for kernel wrapper that wraps two kernel functions.
Parameters: - wrapped1 – first kernel function
- wrapped2 – second kernel function
- class Orange.classification.svm.kernels.RBFKernelWrapper(wrapped, gamma=0.5)¶
A Kernel wrapper that wraps the given function into RBF
Parameters: - wrapped – a kernel function
- gamma (double) – the gamma of the RBF
- __call__(inst1, inst2)¶
Return
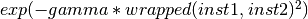
- class Orange.classification.svm.kernels.PolyKernelWrapper(wrapped, degree=3)¶
Polynomial kernel wrapper.
Parameters: - wrapped – a kernel function
- degree (float) – degree of the polynomial
- __call__(inst1, inst2)¶
Return
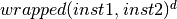
- class Orange.classification.svm.kernels.AdditionKernelWrapper(wrapped1, wrapped2)¶
Addition kernel wrapper.
Parameters: - wrapped1 – first kernel function
- wrapped2 – second kernel function
- __call__(inst1, inst2)¶
Return
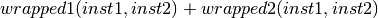
- class Orange.classification.svm.kernels.MultiplicationKernelWrapper(wrapped1, wrapped2)¶
Multiplication kernel wrapper.
Parameters: - wrapped1 – first kernel function
- wrapped2 – second kernel function
- __call__(inst1, inst2)¶
Return
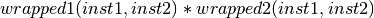
- class Orange.classification.svm.kernels.CompositeKernelWrapper(wrapped1, wrapped2, l=0.5)¶
Composite kernel wrapper.
Parameters: - wrapped1 – first kernel function
- wrapped2 – second kernel function
- l (double) – coefficient
- __call__(inst1, inst2)¶
Return
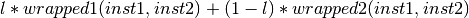
- class Orange.classification.svm.kernels.SparseLinKernel¶
- __call__(inst1, inst2)¶
Compute a linear kernel function using the instances’ meta attributes. The meta attributes’ values must be floats.
Example:
from Orange import data
from Orange import evaluation
from Orange.classification.svm import SVMLearner, kernels
from Orange.distance import Euclidean
from Orange.distance import Hamming
iris = data.Table("iris.tab")
l1 = SVMLearner()
l1.kernel_func = kernels.RBFKernelWrapper(Euclidean(iris), gamma=0.5)
l1.kernel_type = SVMLearner.Custom
l1.probability = True
c1 = l1(iris)
l1.name = "SVM - RBF(Euclidean)"
l2 = SVMLearner()
l2.kernel_func = kernels.RBFKernelWrapper(Hamming(iris), gamma=0.5)
l2.kernel_type = SVMLearner.Custom
l2.probability = True
c2 = l2(iris)
l2.name = "SVM - RBF(Hamming)"
l3 = SVMLearner()
l3.kernel_func = kernels.CompositeKernelWrapper(
kernels.RBFKernelWrapper(Euclidean(iris), gamma=0.5),
kernels.RBFKernelWrapper(Hamming(iris), gamma=0.5), l=0.5)
l3.kernel_type = SVMLearner.Custom
l3.probability = True
c3 = l1(iris)
l3.name = "SVM - Composite"
tests = evaluation.testing.cross_validation([l1, l2, l3], iris, folds=5)
[ca1, ca2, ca3] = evaluation.scoring.CA(tests)
print l1.name, "CA: %.2f" % ca1
print l2.name, "CA: %.2f" % ca2
print l3.name, "CA: %.2f" % ca3
